1 Basics of R Programming
This module is ideal for individuals who have little to no experience with R, and those who would like a reminder. It contains the basics for getting started with R, beginning with how to install R on your computer, and understanding the key aspects of the software. Lastly, Module 1 gives an introduction to using R, including information on the basic operations, functions, and data structures.
1.1 Overview of R
R is a free, open source statistical programming language and environment used for statistical computing and has a wide range of graphical facilities for data analysis. Its uses also include (but not limited to) the following:
- Data manipulation - e.g. extract, clean, store, process, …
- Calculations (simple and complex) - e.g. addition, subtraction, …
- Data visualisation - e.g. charts, line graphs, maps, exploratory analysis, …
- Statistical and data analysis - e.g. regression, t-tests, correlation, …
- Machine and deep learning - e.g. modelling, cross-validation, descriptive statistics, …
- Reporting and creating documents (using Markdown and LaTeX) - e.g. reports, presentations, reproducible research, …
- Mapping (Geographic Information System, GIS) - e.g. interactive visualisation, population size estimation,…
Getting started with R
Downloading R and RStudio
R can be downloaded on Windows, Mac and Linux from R-Project.org. Select the link to download for the correct operating system for your computer.
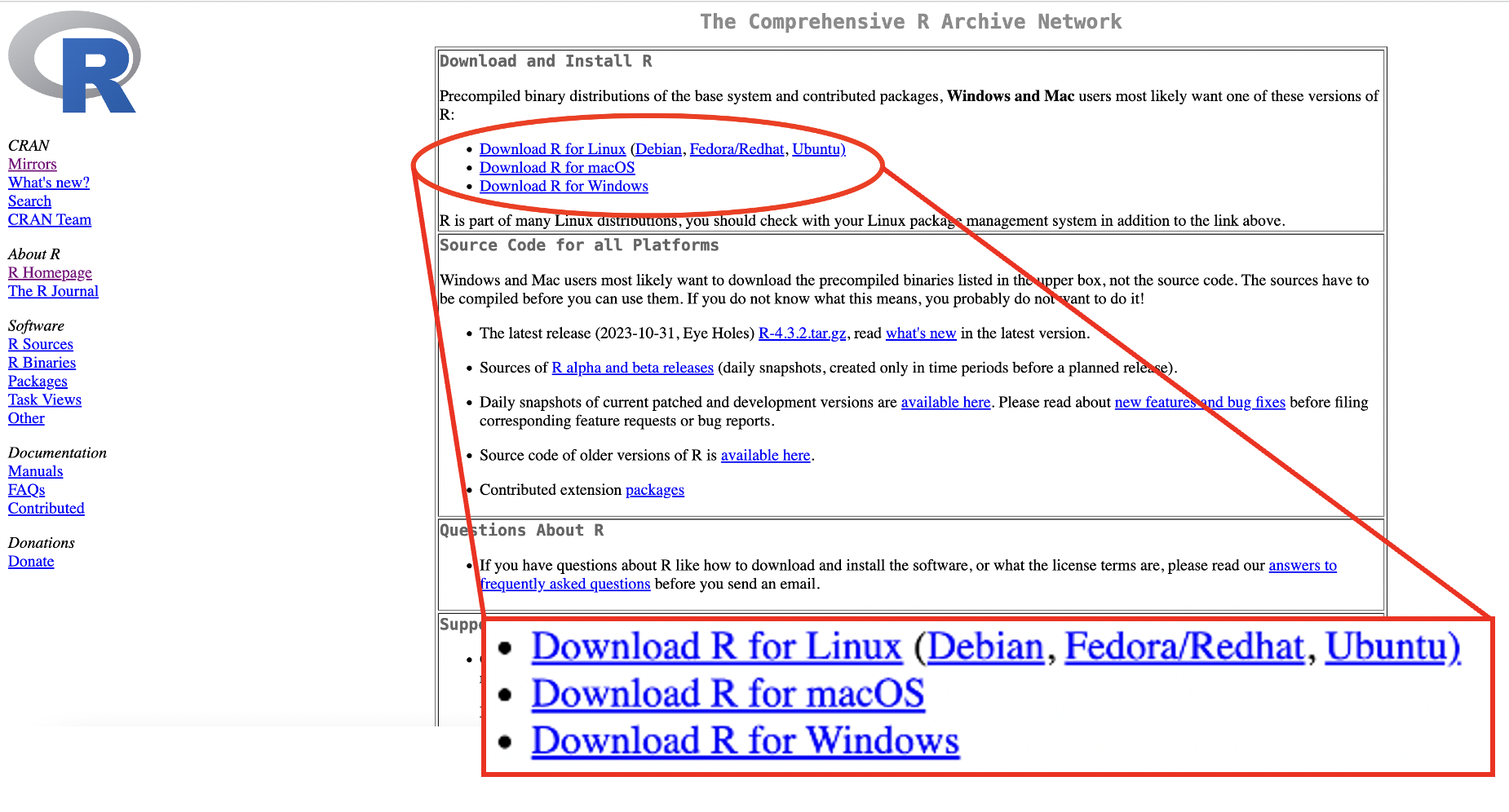
Figure 1.1: R download webpage
For Windows, click “install R for the first time”.
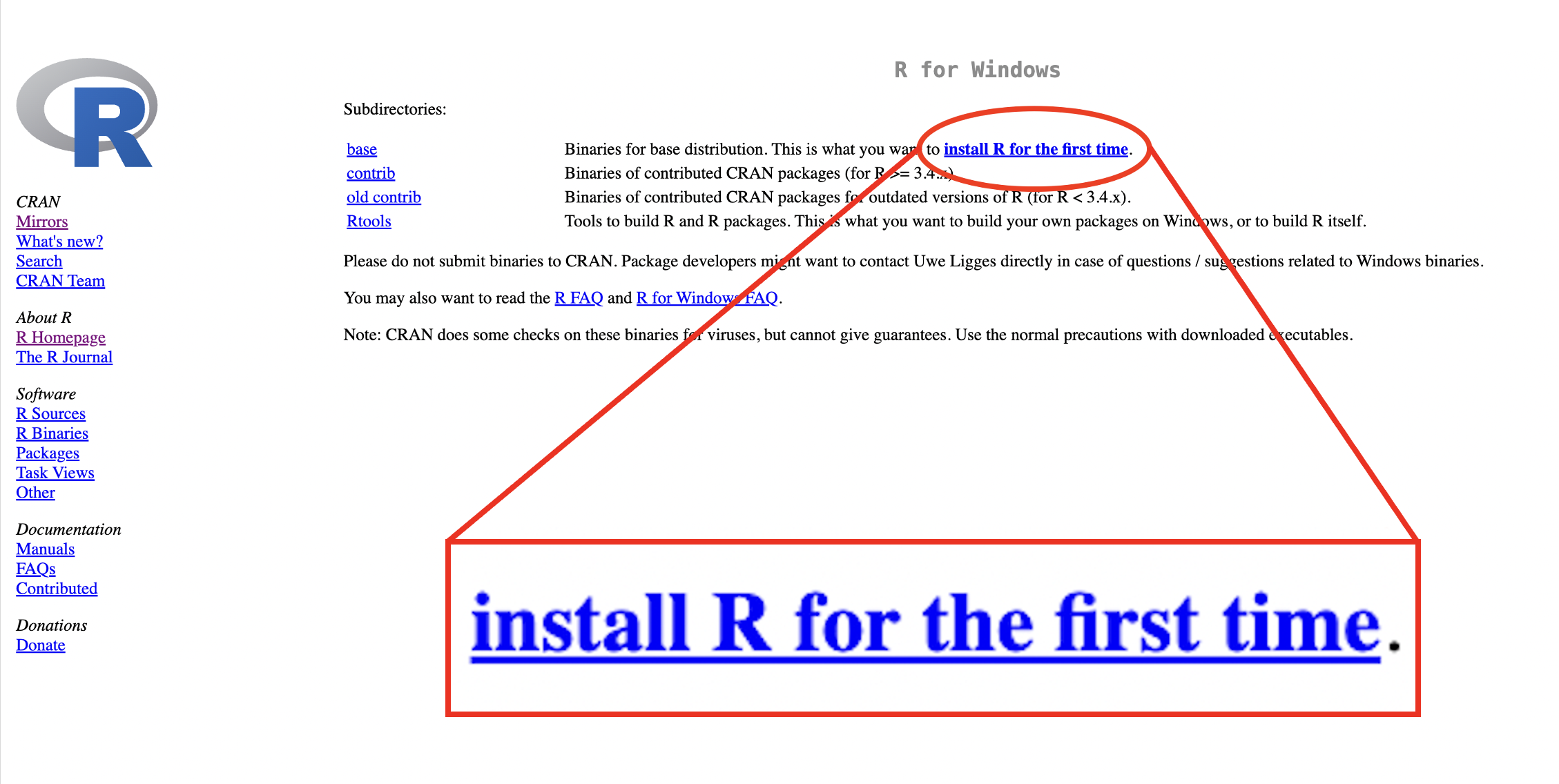
Figure 1.2: R download webpage: Windows
For macOS, click the correct download package for your Mac’s software version.
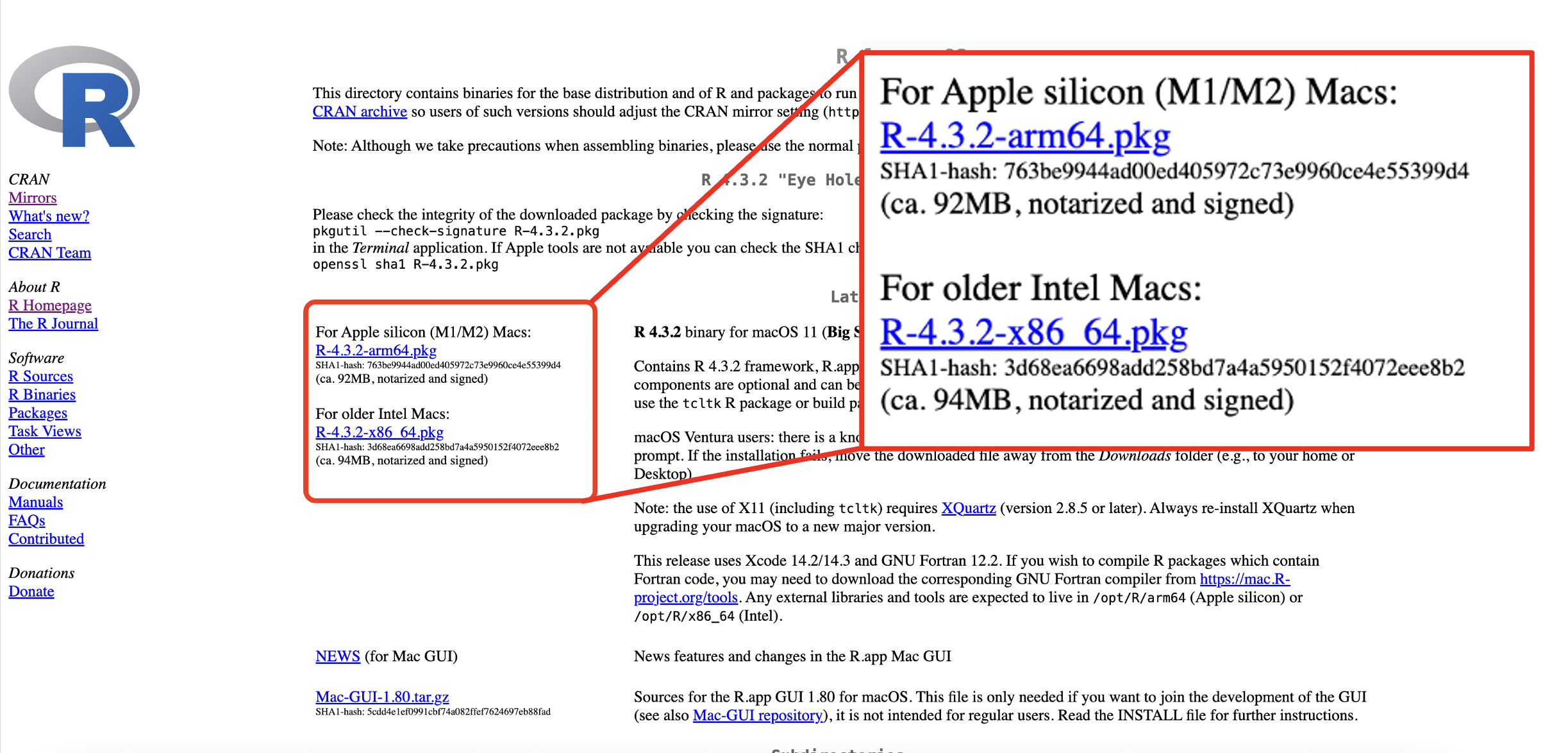
Figure 1.3: R download webpage: macOS
R does not have a particularly user-friendly graphical user interface (GUI). RStudio is a capable and free GUI add-on for R with improved functionality and usability. It can be downloaded from Postit.co. The webpage for downloading RStudio detects whether the operating system is Windows or macOS (or Linux), so the correct operating system does not need to be selected.
For Windows users, the webpage will look like the image below.
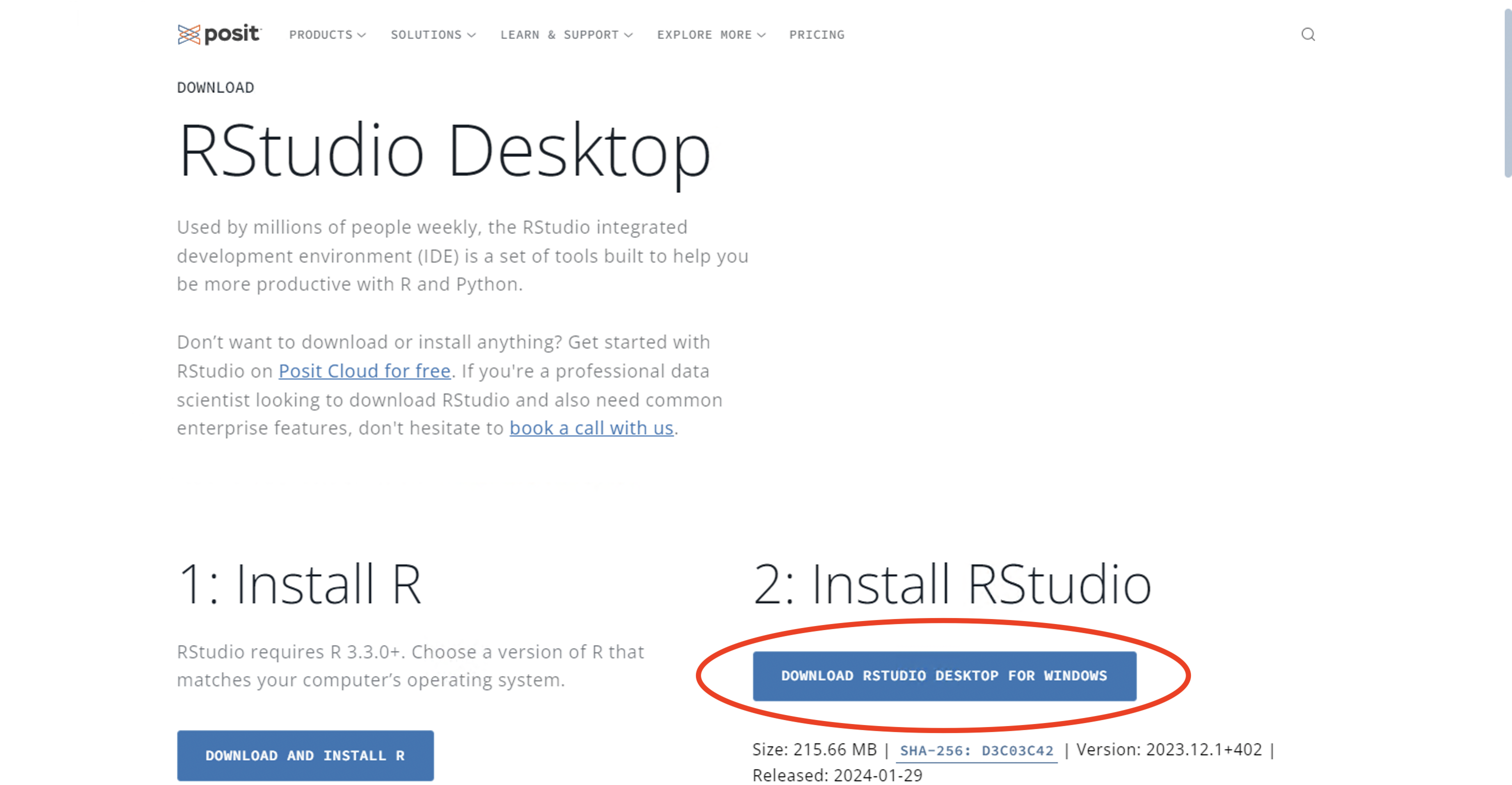
Figure 1.4: RStudio download webpage: Windows
For macOS users, the webpage will look like the image below.
Figure 1.5: RStudio download webpage: macOS
Once “Download RStudio desktop” has been selected, the preferred version of RStudio can be selected, e.g. 2023.03.1+446, and the installation process can be followed once the download is complete.
To check both R and RStudio are installed on the computer, search for “R” and “RStudio” in the search bar of the computer. Once both are installed, the process is complete and they can be used.
Note: RStudio is just one example of R GUI, others are available such as Positron.
Features of the R GUI environment
When opening R, the user interface should have a window open for the console, where calculations can be made and results will be printed. It should look something like the image below.
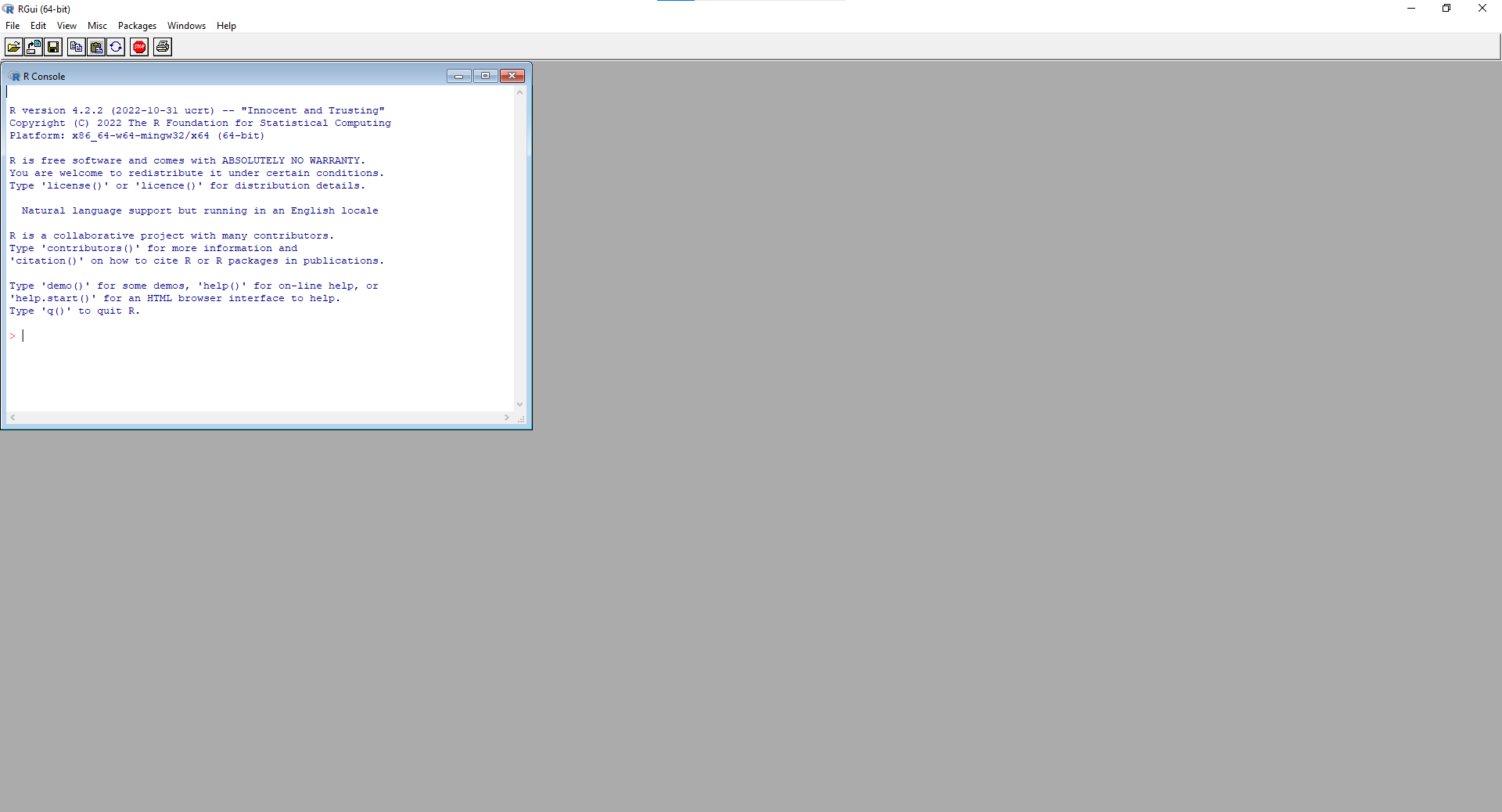
Figure 1.6: R GUI upon opening
Additional windows can be opened, such as windows for scripts to code in and windows to display plots, seen in the image below.
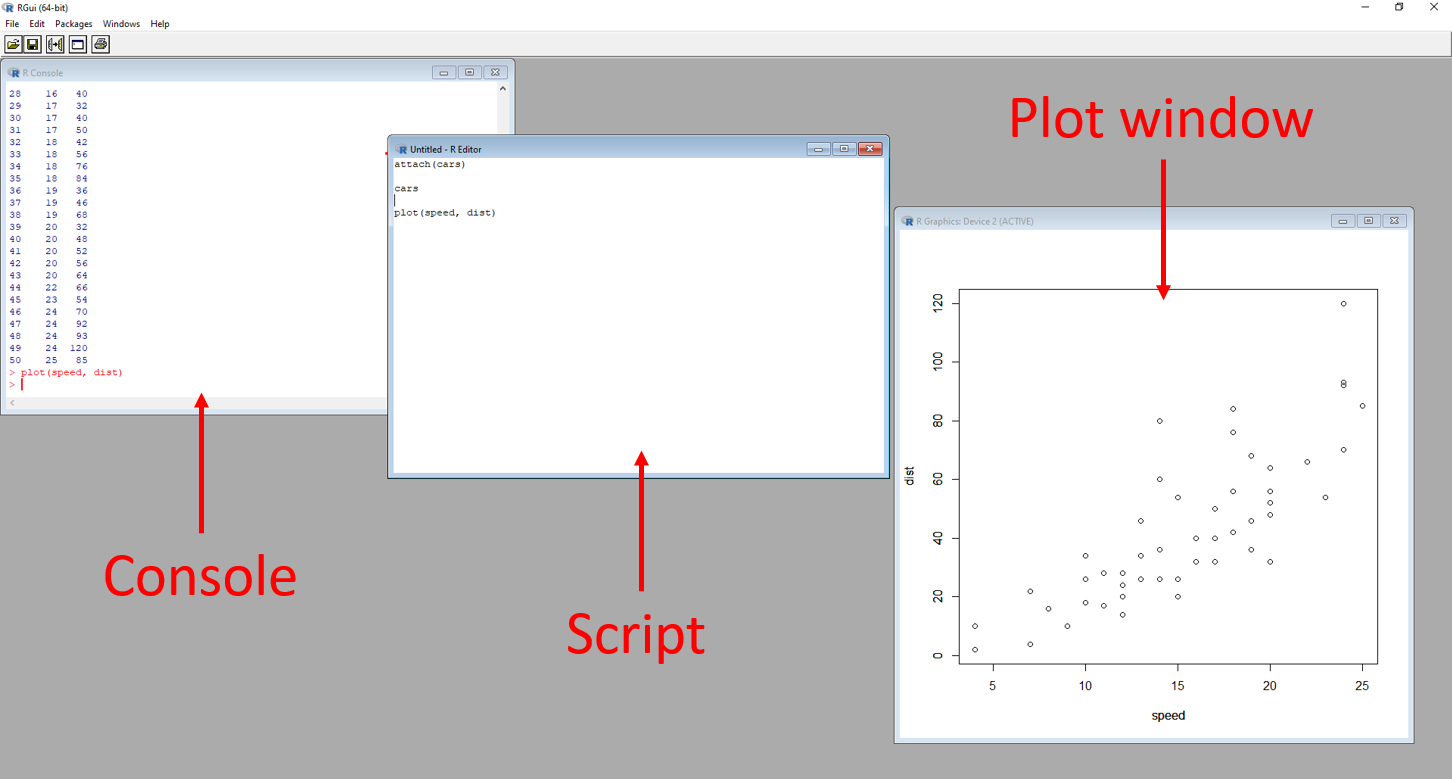
Figure 1.7: Annotated R GUI
Features of the RStudio environment
The user interface for RStudio is more efficient, with designated panes for displaying different information all at once in a much more convenient way.
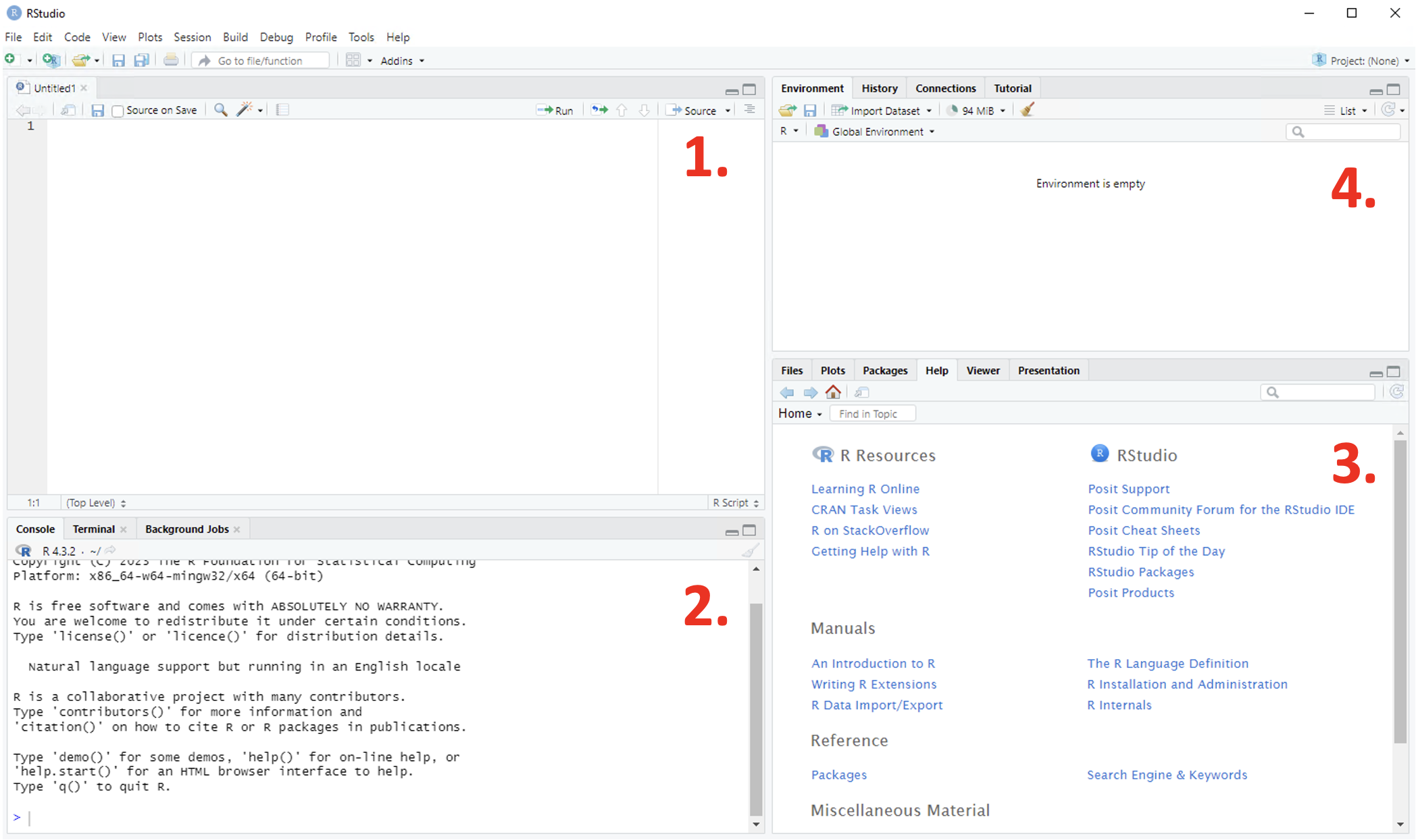
Figure 1.8: Annotated RStudio GUI
There are four panes as seen in the figure below:
- Script - this pane is only present once a script has been created
- Console - where numerical output will be returned
- Viewer - computer files, plots, packages, help files, reports and presentations can be viewed here
- Environment - any objects that have been created are present in the environment and can be cleared by clicking on the ‘broom’ icon toward the centre of the toolbar
See the image below for assistance in navigating the key aspects of the RStudio GUI.
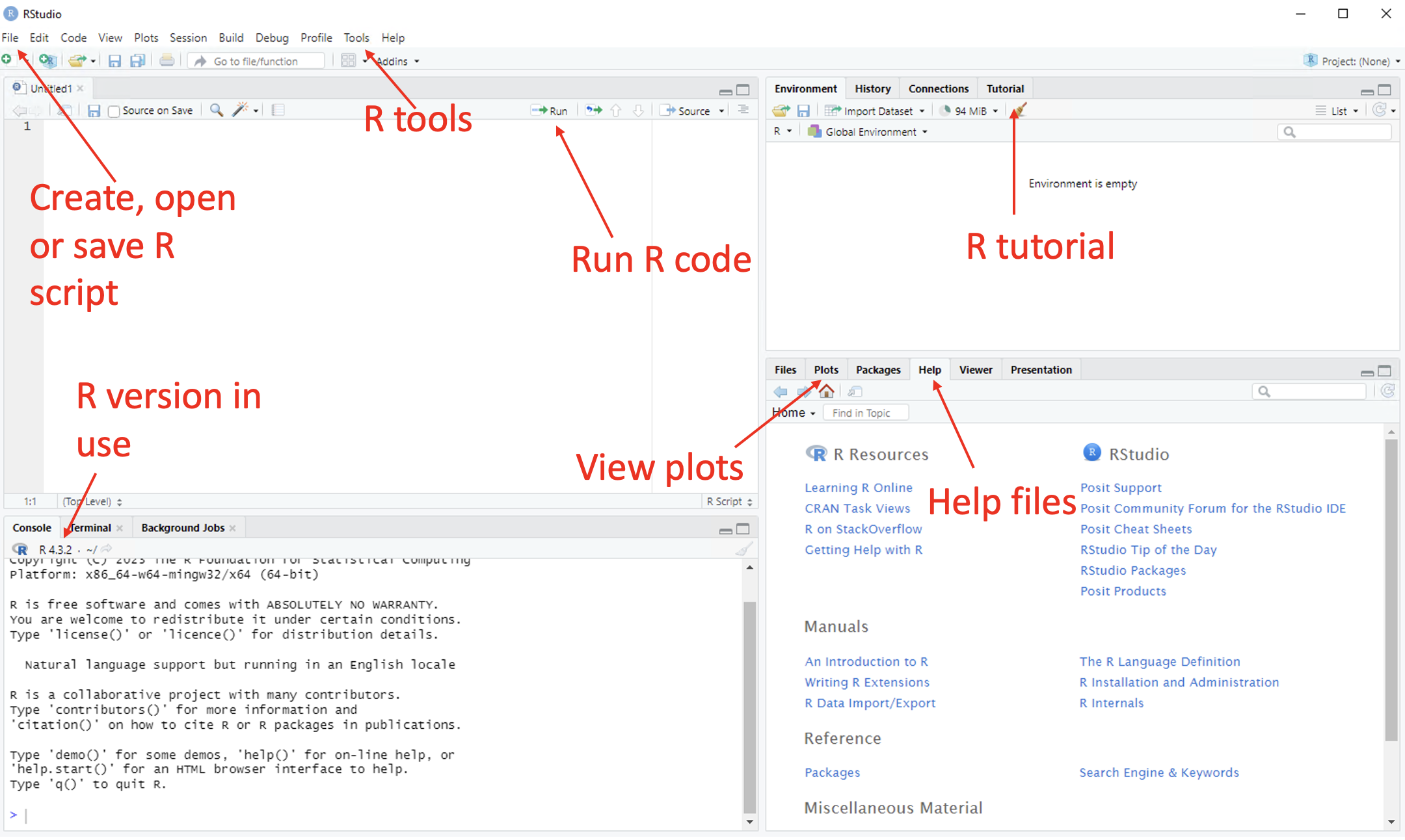
Figure 1.9: Annotated RStudio GUI
Creating, opening and saving R scripts
Whilst commands can be typed directly into the console, it is difficult to keep record of what has been run to ensure the code is reproducible and inconvenient when typographical mistakes are made. Utilising a script solves these issues, with the ability to make changes to and save the code written.
In RStudio, to open a new script, use the main toolbar and follow File > New File > R Script and pane 1 will appear. If a script is already present, it will open the new script in a new tab in the pane.
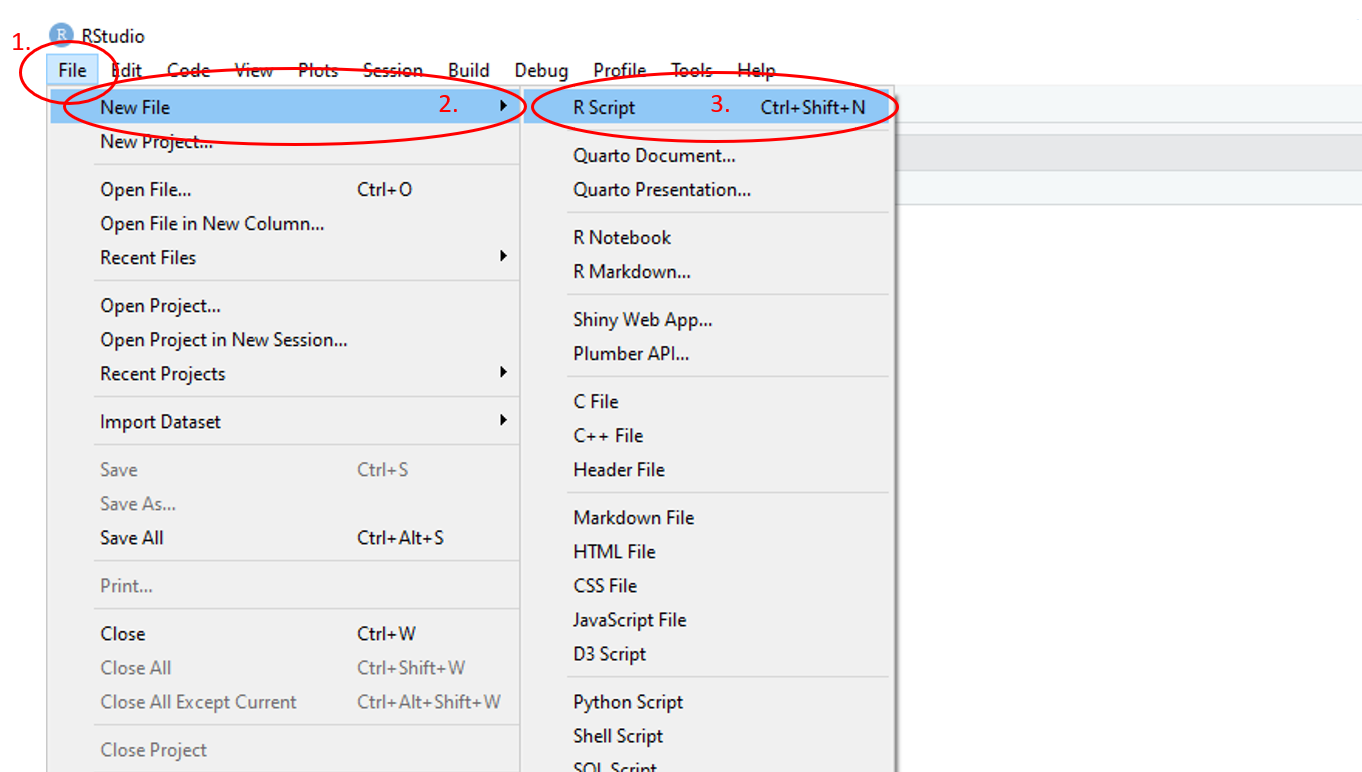
Figure 1.10: Annotated RStudio GUI: New script
To run the code from a script, highlight the desired line(s) to run and click Run on the script pane at the top right. Alternatively, keyboard short cuts can be used to run the code, press Control + Enter on Windows or Command + Enter on Mac. If code is run without highlighting any lines, only the current line (where the cursor is) will be run.
To open an existing R script, use the main toolbar and follow File > Open File then navigate to the desired R script in the files pop-up window.
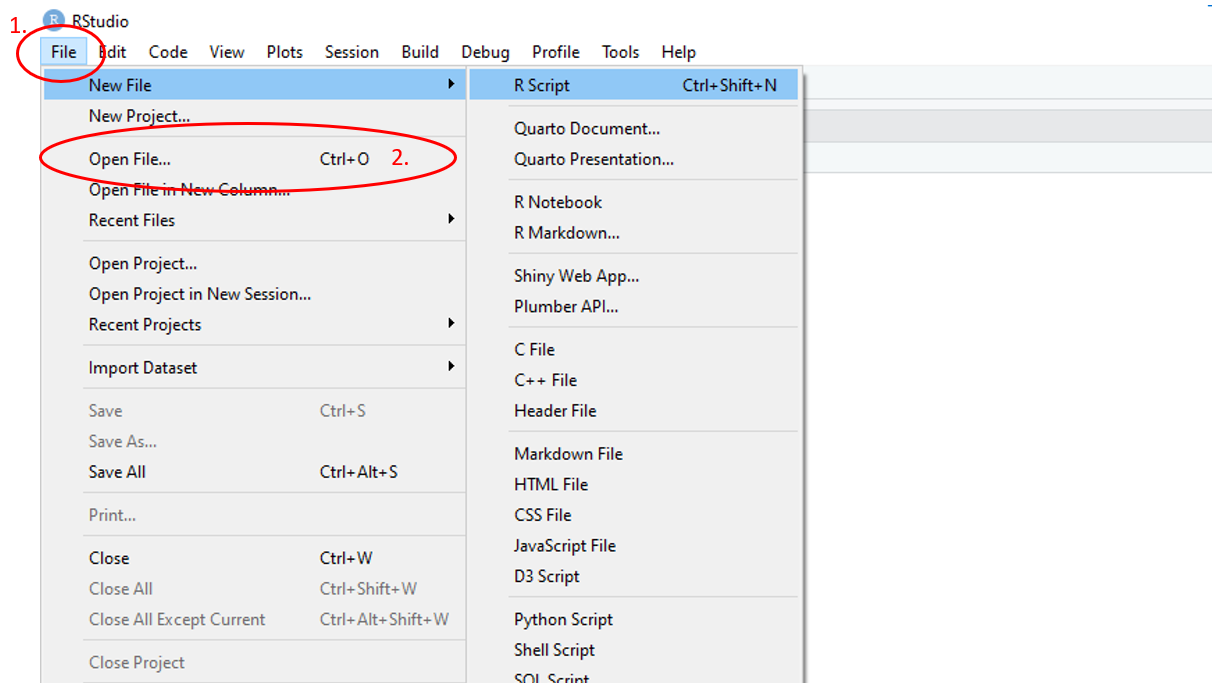
Figure 1.11: Annotated RStudio GUI: Open script
To save an R script, use the main toolbar and follow File > Save As and navigate to the desired location in the pop-up file directory window and name the script. To save updated versions of the script, use the main toolbar and follow File > Save
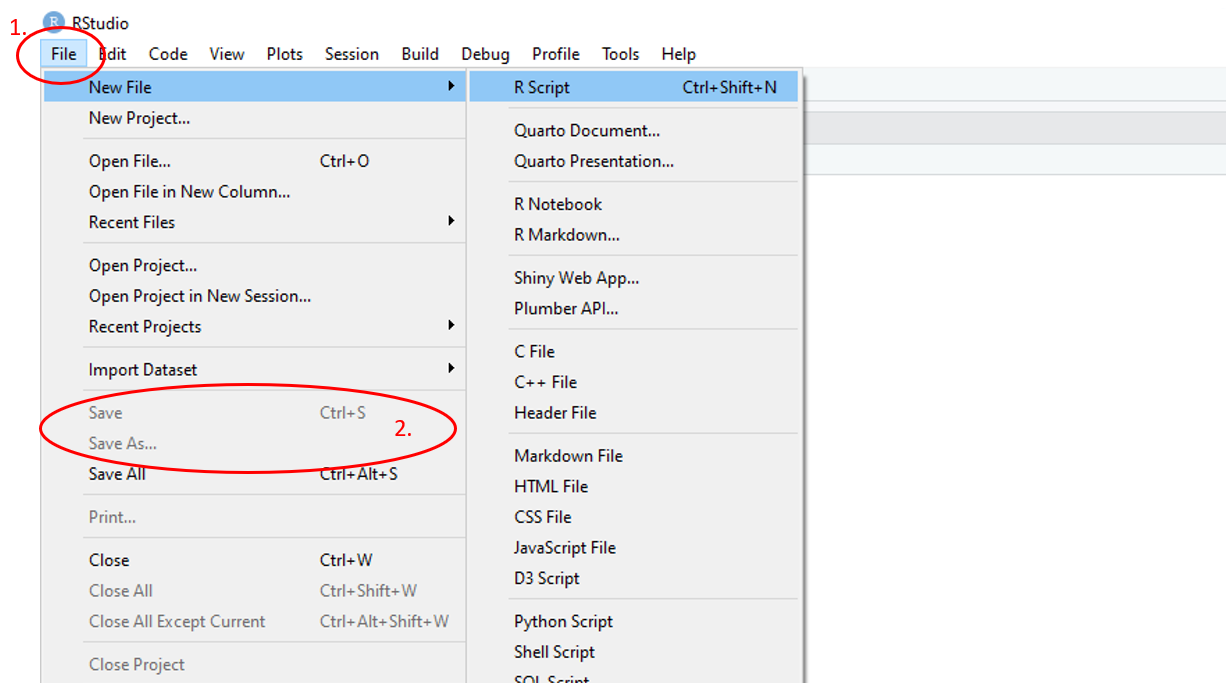
Figure 1.12: Annotated RStudio GUI: Save script
1.1.1 Getting and setting the working directory
The working directory is the folder on the computer where RStudio will access data and files from and/or save data to without any extra commands or steps. To avoid data and files being saved in an unknown location as well as to allow RStudio to access the data, it is important to know the working directory of your R session.
To check the location of the working directory, use the function getwd(). In the console, the working directory file path name will be printed.
To set the working directory, you use the function setwd(“location of your data folder”) and include the file path name in the parentheses.
Alternatively, the main toolbar menus can be used to set the working directory following the steps below, also demonstrated in the figure below:
- Click on “Session”
- Navigate to “Set Working Directory”
- Navigate to “Choose Working Directory”
- Select in your computer where you would like your R session files saved
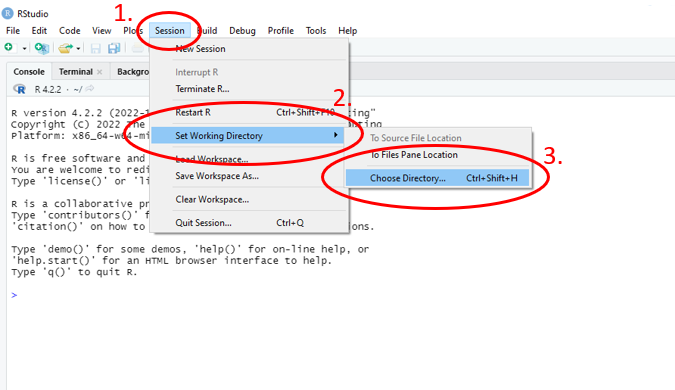
Figure 1.13: Annotated RStudio GUI: Set working directory
1.1.2 Creating, opening and saving R projects
When working with files in R, it is imperative that the working directory is set at the start of each script in order for R to be able to access the files. However, when working on different computers or working collaboratively with others, the file path will change leading to reproducibility issues.
An R Project is essentially a reproducible working directory, preventing the need to use the function setwd() in each script through having all of your work in a self-contained folder with a designated .Rproj file which can be opened and run seamlessly by anybody who has access.
To create an R project, use the main toolbar and follow the path File > New Project… > New Directory > New Project, give your directory a name and then click on Create Project.
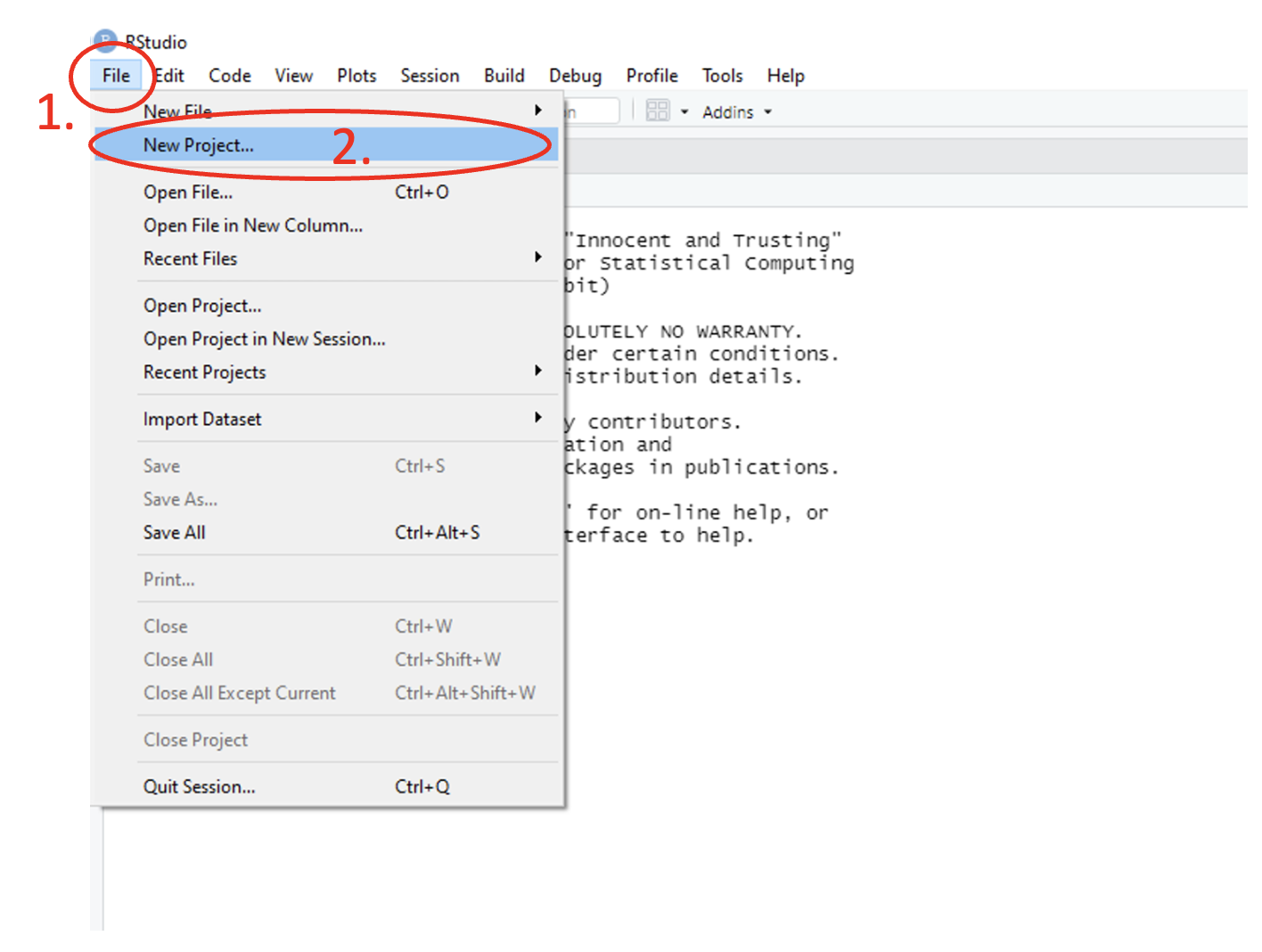
Figure 1.14: Annotated RStudio GUI: Create project
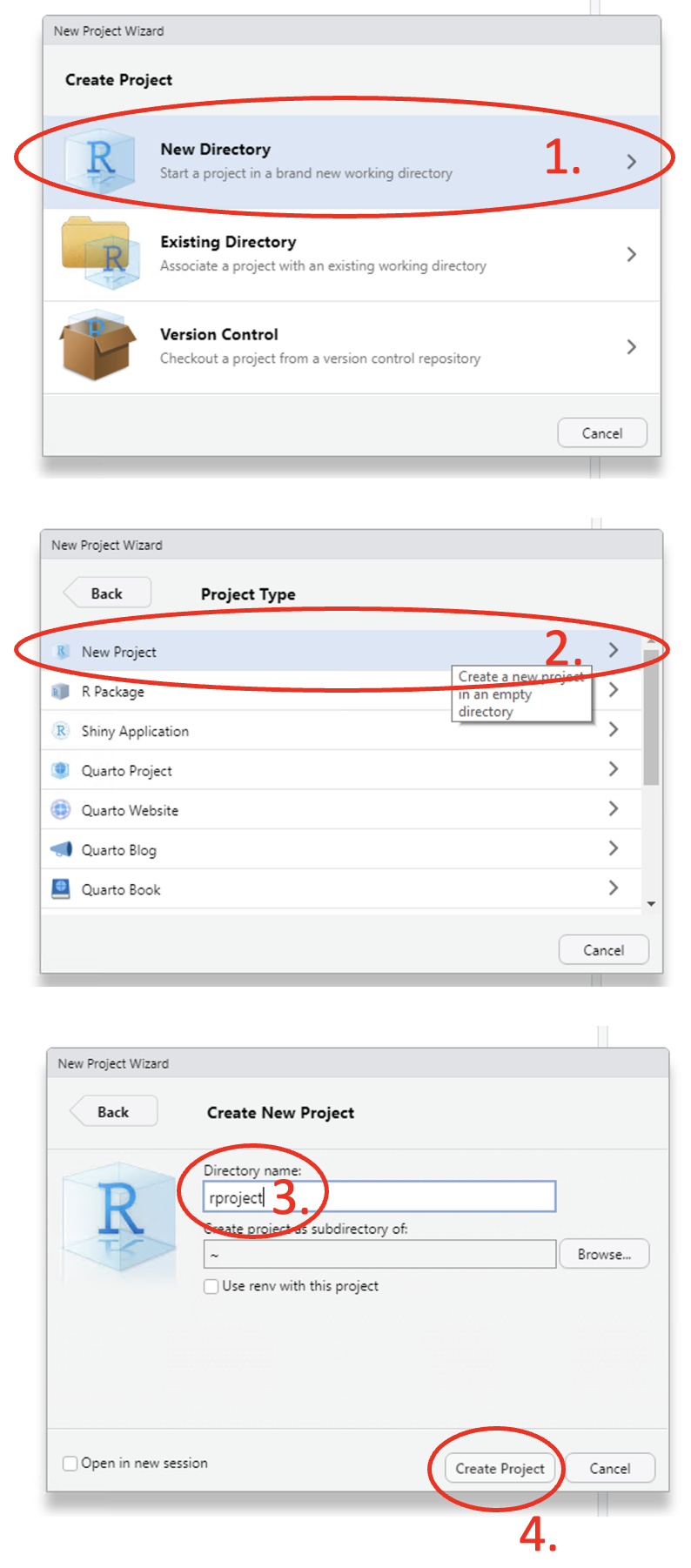
Figure 1.15: Annotated RStudio GUI: Create project
To open an existing R project, use the main toolbar and follow the path File > Open Project… > and select the desired project from the pop-up file directory window.
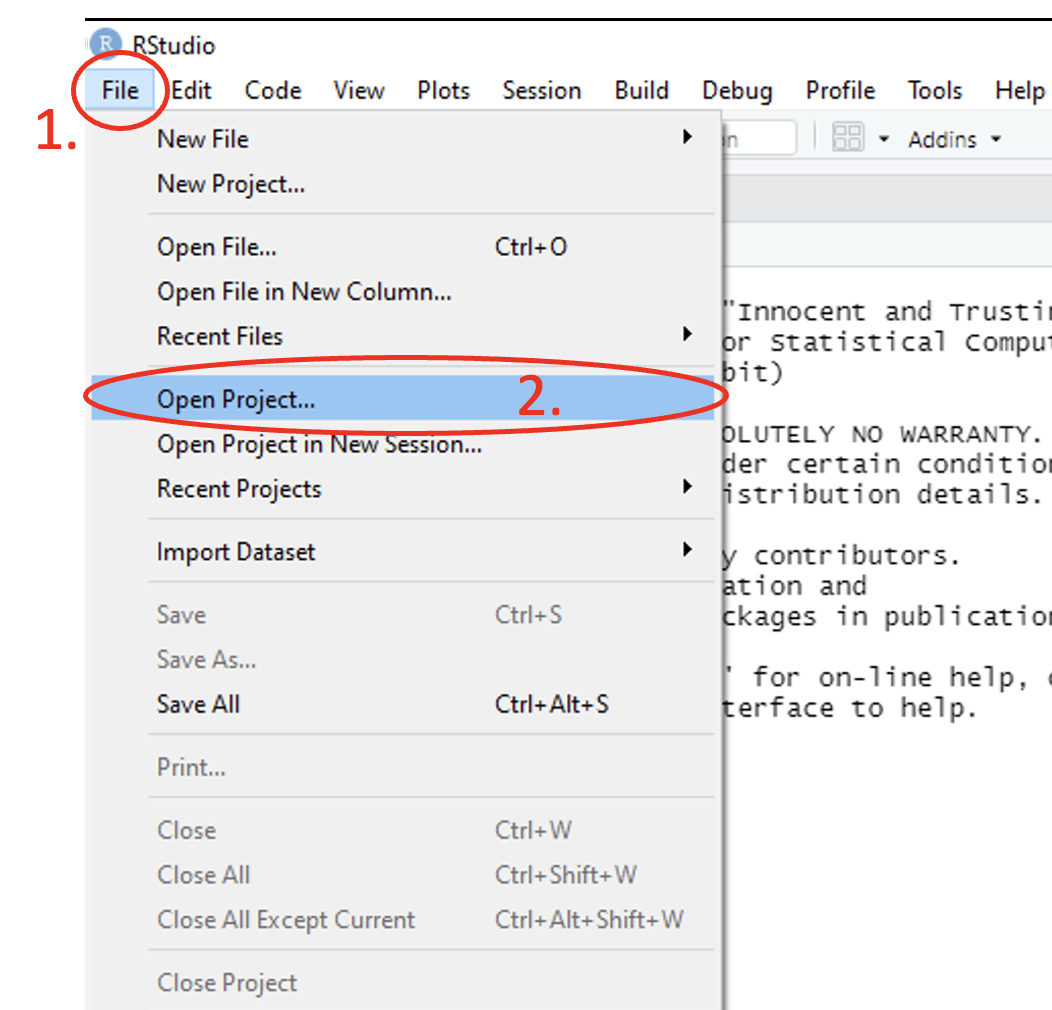
Figure 1.16: Annotated RStudio GUI: Open project
To save an R project, the same method is used as for R scripts, using the main toolbar and following the path File > Save As and navigate to the desired location in the pop-up file directory window and name the project To save updated versions of the project, use the main toolbar and follow File > Save.
1.1.3 Getting help in R
There are inbuilt facilities which provide information to help about any specific command or package in R.
The first method to get help in R is to run lines of code. For example, to get information about ‘mean’, run any of the following
help(mean): displays help in pane 3?mean: displays help in pane 3help.start(mean): displays help as html??mean: displays help as html
Alternatively, use the toolbar of pane 3, navigate to the Help tab and use the search bar to look for the command.
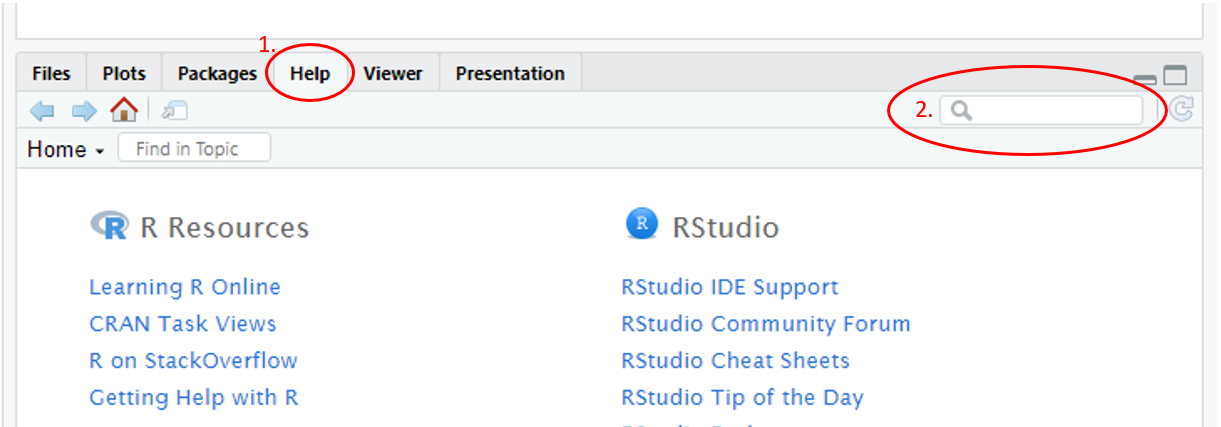
Figure 1.17: Annotated RStudio GUI: Help files
The help files for R functions and commands contain information on the following sections:
- Description: A brief description on what the function(s) does and its usage
- Usage: How to use the function(s) with what arguments need to be supplied and any default values of arguments
- Arguments: Explanation on what the arguments are
- Details: More in-depth information on the background of the function(s)
- Value: Values that the function(s) returns
- Source: What the function(s) is based on
- References: Bibliography used for creating function(s)
- See Also: Links to similar functions that may be of use
- Examples: Examples on how to use the function(s)
If the information provided in the help files is insufficient, Stack Overflow is a great website where individuals can ask and answer questions. There are over 300,000 questions already asked on there so it is likely that the answer to your question has already been asked.
For some aspects of R, there are ‘Cheat Sheets’ available, posters which provide the essential, key information on the topic. To access this help, use the main toolbar and follow Help > Cheat Sheets > and select the file which matches your needs best. If the cheat sheet you require is not provided in the list, more options can be found by selecting Browse Cheat Sheets….
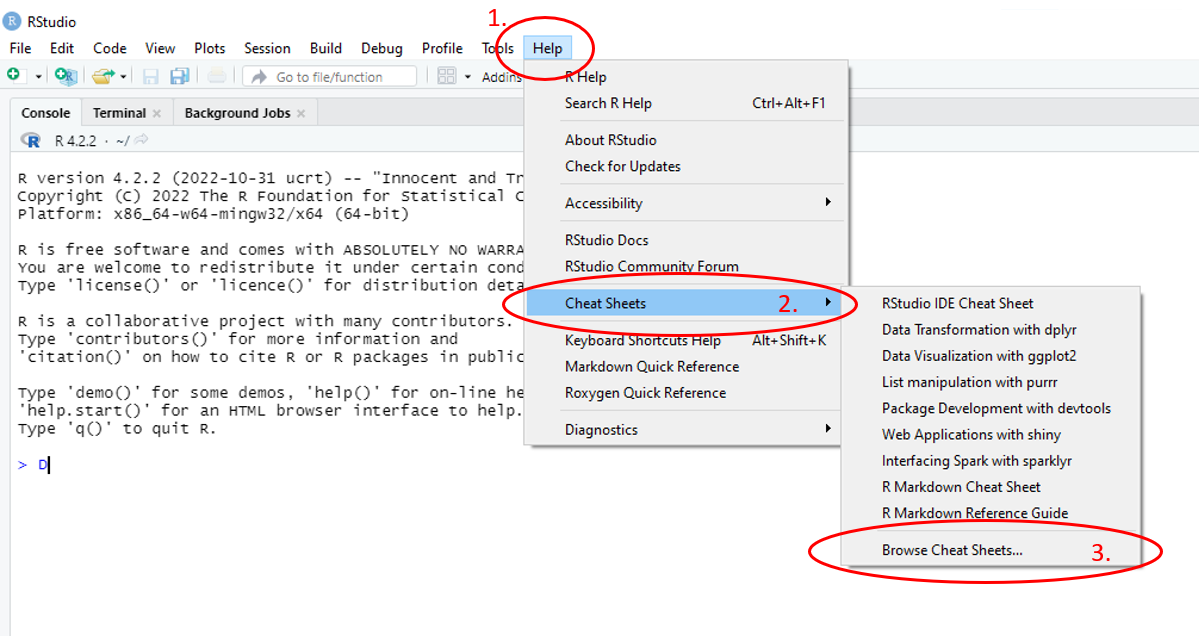
Figure 1.18: Annotated RStudio GUI: Help files
1.1.4 R coding best practices
1.1.4.1 Commenting
Including comments describing the code is good practice as it ensures that the code can be understood by both yourself in the future and others that the code is shared with. Comments can be added into the code by using the hash key, #, and stops the line being commented from running with the rest of the code.
#adding a comment with a hash tag ensures that yourself and others can understand
#the code quickly if returned to after some time.
X <- c(2, 3, 5, 7, 11, 13, 17, 19, 23, 29) #X contains prime numbers in orderAdditionally, the hash can be used to create sections within scripts. To create a section, use at least 3 in a row without spaces in between (###). This is good practice for keeping your scripts organised.
1.1.4.2 Naming conventions
It is not only important to ensure anything in your code that is assigned a name is clear and identifiable, but also that the name chosen does not conflict with something that already has assignment by R (for example, c is already assigned to a function in R, used for combining values into vectors or lists). Some names are reserved by R, such as the names of existing functions. These names should not be used as your code will overwrite the existing code and the functions/values will no longer work.
To ensure that you are not assigning names that are not already taken, you can run the desired name and if it returns any code that you did not write, then R has already reserved that name. However, if it returns an error message, then the name is available for you to use.
#check if the name "abs" is available
abs #"abs" is not available as it returns code for an existing function## function (x) .Primitive("abs")#check if "absolute" is available instead
absolute #returns error message, "absolute" is free to use## Error: object 'absolute' not found## function (...) .Primitive("c")## [1] 3.1415931.1.4.3 Spacing
R ignores spaces provided that they are not in the middle of a command name or operator. As a result, R treats the following as identical lines of code.
However, if you include the space in the middle of the operator, R will not return the result, but FALSE instead.
## [1] FALSER also ignores indentations at the start of a line, so spaces at the start of a line of code will appear the same as if there are no spaces. This is different to Python.
1.2 Data types and key components
There are six common data types:
- Boolean: logical statements e.g.
TRUE,FALSE,T,F - Character: letters or non-numbers e.g. “alphabets”, “names”, …
- Factor: non-numbers (characters) with levels, both ordered and unordered, with a predefined, finite number of values e.g. marital status field only allowing the options of: “single”, “separated”, “married”, “widowed/widower” or “divorced”
- Integer: whole numbers e.g. 1L, 2L, 100L, …
- Numeric: 1 and any number with decimals e.g. 1, 1.5, 8.9, …
- Complex: complex numbers with real and imaginary parts e.g. 3+4i, 1-I, …
To check the data type in R, use the class() function.
There are three key components in R:
- Objects: used to store values
- Functions: allow for manipulations on objects to be performed
- Operators: used to create interactions between objects or between an object and a function
1.2.1 Basic operations in R
1.2.1.1 Arithmetic operators
| Operator | Description |
|---|---|
+ |
addition |
- |
subtraction |
* |
multiplication |
/ |
division |
^ or ** |
exponentiation |
%% |
modulus of a number |
%/% |
integer division |
%*% |
matrix multiplication |
1.3 Key functions in R
Functions are always indicated with a name followed by (), and anything inside the parentheses is an argument, or option that is passed. Some arguments have a default value, meaning that if no value for that argument is given in the code, the default value will be used.
The following table contains some of the more commonly used functions that are available in base R, with a description of what they are used for. Examples on how to use many of these functions are seen later in the module.
| Function | Description |
|---|---|
log() |
logarithm |
exp() |
exponential |
abs() |
absolute value |
seq() |
sequence of values |
sqrt() |
square root |
sd() |
standard deviation |
var() |
variance |
median() |
median |
mean() |
mean |
quantile() |
quantiles |
sum() |
add all elements together |
diff() |
difference |
min() |
minimum value/ element |
max() |
maximum value/ element |
range() |
gives smallest & largest numbers |
table() |
tabulation |
length() |
length of the vector |
summary() |
summarise the entire vector |
c() |
combine values into list or vector |
cbind() |
combine vectors, matrices or data frame by column |
rbind() |
combine vectors, matrices or data frame by row |
which() |
gives the TRUE indices of a logical object |
round() |
rounds the input to the specified accuracy |
paste() |
link vectors after converting to characters |
paste0() |
pastes everything as given |
cat() |
converts arguments to character strings |
print() |
outputs the object(s) given |
Exercise: Which of the following will give a different output from the other 3?
Hint: Look at the help file for the log() function.
log(x=6, base=4)log(4, 6)log(base=4, x=6)log(6, 4)
1.4 Data structures
There are five data structures:
- Vector: one dimensional where all elements must be of the same type, either numeric, character or factor
- Matrix: two dimensional where data elements of the same data type are stored in rows and columns
- Array: a data structure that contains multiple matrices where the elements are arranged sequentially
- Data frame: two dimensional and can contain vectors of different data structures and can combine different variables
- List: a flexible data structure that can contain all other data structures
1.4.1 Vectors
A vector can be a sequence of numbers, characters, logical, complex or a combination, and can be created in numerous ways.
One way of creating a vector is to use the function c() which combines the values input as arguments into a vector, providing that each value is separated with a comma.
## [1] 1 2 5 3 7If creating a vector that is a string of characters, it is imperative that each character is separated is enclosed by either ' or ".
## [1] "Northern" "Northeastern" "Western" "Central" "Eastern" "Southern"A vector can be created to contain a combination of both numbers as strings.
## [1] "4" "oranges" "5" "apples" "12" "carrots"Factors are related to vectors, and in R, factors are stored as integer vectors, where there are a finite number of categories and each level (or label) is a character. These can be created with the factor() function, inputting the vector you wish to convert to a factor as the argument.
#create factor for marital status
marital_status <- factor(c("single", "separated", "married", "widowed/widower",
"divorced", "married", "widowed/widower", "single",
"single", "married"))
#transform Regions to factor
Regions_fac <- factor(Regions)To access the levels of the factor, you can either just print the variable itself or use the levels() function, inputting the factor variable as the argument.
## [1] single separated married widowed/widower divorced married
## [7] widowed/widower single single married
## Levels: divorced married separated single widowed/widower## [1] "Central" "Eastern" "Northeastern" "Northern" "Southern" "Western"The structure of the factor can be accessed with the str() function, where the output shows the different levels and indicates which level each of the elements of the variable are.
## Factor w/ 5 levels "divorced","married",..: 4 3 2 5 1 2 5 4 4 2Note: the data.frame() function explored later in this module automatically converts character vectors into factors if the argument stringsAsFactors=FALSE is not passed.
Another way to create a vector is to use a colon : which generates a regular sequence. a:b creates a sequence of numbers from a to b in increments/steps of either +1 or -1.
## [1] 1 2 3 4 5 6 7 8 9 10## [1] 20 19 18 17 16 15 14 13 12 11 10#'a' and 'b' do not need to be integers:
#vector of a sequence from 5.5 to 15.5, going up in increments of 1
5.5:15.5 ## [1] 5.5 6.5 7.5 8.5 9.5 10.5 11.5 12.5 13.5 14.5 15.5Also to create a vector, the function seq() in R can be used, with the following arguments:
from: the starting value (default=1)to: the end value (default=1)by: a numeric value indicating the size of each increment/step (default= (to-from)/(length.out-1))length.out: a non-negative numeric value indicating the desired length of the sequence
## [1] 1 3 5 7 9 11 13 15 17 19 21 23 25 27 29 31 33 35 37 39 41 43 45 47 49 51#vector of a sequence from 1 to 50, with the sequence of length 20
seq(from = 1, to = 50, length.out = 20) ## [1] 1.000000 3.578947 6.157895 8.736842 11.315789 13.894737 16.473684 19.052632 21.631579 24.210526
## [11] 26.789474 29.368421 31.947368 34.526316 37.105263 39.684211 42.263158 44.842105 47.421053 50.000000The function rep() which replicates the value(s) input can also be used to create a vector in R, using the following arguments:
x: a number, vector or a factor which you want to replicatetimes: a non-negative integer or integer-valued vector giving the number of times to repeatxeach: a non-negative integer giving the number of times to repeat each element inxin order/consecutivelylength.out: a non-negative integer to give the value of the desired length of the vector
## [1] 1 1 1 1 1 1 1 1 1 1#vector of a sequence from 1 to 10, going up in increments of 1, repeated twice
rep(x = 1:10, times = 2) ## [1] 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10#vector of a sequence from 1 to 10, going up in increments of 1, where each
#element is repeated twice consecutively
rep(x = 1:10, each = 2) ## [1] 1 1 2 2 3 3 4 4 5 5 6 6 7 7 8 8 9 9 10 10#vector of a sequence from 1 to 10, going up in increments of 1, repeated until
#the length of the vector is 15
rep(x = 1:10, length.out = 15) ## [1] 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5Exercise: Create a vector that goes from 5 to 25 by increments of 1.
A combination of the different vector construction methods can be used at once, for example, the combine function c() can be used within the other functions and can also be used to combine existing vectors.
vec1 <- seq(from = 10, to = 100, by = 10)
vec2 <- rep(x = 15:10, each = 2)
#combine vectors vec1 and vec2
vec3 <- c(vec1, vec2)
vec3## [1] 10 20 30 40 50 60 70 80 90 100 15 15 14 14 13 13 12 12 11 11 10 10Exercise: Create another vector that goes from 5 to 25 by increments of 1, but using a different method. Call this vector V1
Elements from vectors can be indexed (selected) using square brackets [].
## [1] 10## [1] 15 14 13## [1] 15 15 14 13 12 12 11 11 10 10#element-wise calculations: multiply the third (3rd) element from vec3 by the
#fourth (4th) element of vec2
vec3[3]*vec2[4] ## [1] 420Exercise: Extract the 5th element of V1.
Through indexing elements from vectors, elements within vectors can be updated.
## [1] "4" "oranges" "5" "apples" "12" "carrots"#update the first (1st) element of mixed_vec from 4 to 6
mixed_vec[1] <- 6
#update the second (2nd) and fourth (4th) elements of mixed_vec from oranges
#and apples to lemons and bananas
mixed_vec[c(2, 4)] <- c("lemons", "bananas")
mixed_vec## [1] "6" "lemons" "5" "bananas" "12" "carrots"Exercise: Remove the 7th element from the vector V1 and call this new vector V2.
Vectors can be summarised, inputting the vector into functions different functions.
## [1] 10## [1] 150## [1] 31.81818## [1] 67.4## [1] 67Logical operators can be used with vectors to return TRUE or FALSE for each element depending on the conditions given.
#return TRUE for elements which are both greater than 10 and less than 100, and
#FALSE if both conditions are not met
vec4 > 10 & vec4 < 100 ## [1] TRUE FALSE FALSE FALSE TRUEIndexing used with logical vectors returns the elements in the vector which meet the conditions given.
## [1] 12 84## [1] -1 239The function which() can be used in conjunction with logical operators to return the index/indices of the element(s) which meet the conditions given, rather than the logical statements or values.
## [1] 1 4 5Extensions of the which() function are which.min() and which.max() which respectively give the indices of the elements with the minimum value and the maximum value.
## [1] 3## [1] 4Exercise: How many elements in your vector, V1, are greater than 11?
1.4.2 Matrices
Used for data storage and often faster to work with than a data frame.
Create a matrix using the function matrix() in R, with the following arguments:
data: the data that you want to include in the matrixnrow: the number of rows you want the matrix to havencol: the number of columns you want the matrix to havebyrow: logical, ifFALSE(the default), the matrix fills by columns, ifTRUE, the matrix fills by rowdimnames: (optional,NULLby default) input alistof length 2, containing the row and column names respectively
#example of creating a 4x4 matrix:
mat <- matrix(data = seq(1, 16), nrow = 4, ncol = 4, byrow = TRUE)
mat## [,1] [,2] [,3] [,4]
## [1,] 1 2 3 4
## [2,] 5 6 7 8
## [3,] 9 10 11 12
## [4,] 13 14 15 16#combine two matrices by column:
matrix1 <- matrix(data = 1, nrow = 2, ncol = 2)
matrix2 <- matrix(data = 2, nrow = 2, ncol = 2)
#returns a 2x4 matrix
cbind(matrix1, matrix2) ## [,1] [,2] [,3] [,4]
## [1,] 1 1 2 2
## [2,] 1 1 2 2Matrices can also be created through combining multiple vectors.
#create two vectors of the same length
vec_mat_a <- 1:10
vec_mat_b <- 11:20
#use rbind to create a 2x10 matrix
vec_mat1 <- rbind(vec_mat_a, vec_mat_b)
vec_mat1## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## vec_mat_a 1 2 3 4 5 6 7 8 9 10
## vec_mat_b 11 12 13 14 15 16 17 18 19 20## vec_mat_a vec_mat_b
## [1,] 1 11
## [2,] 2 12
## [3,] 3 13
## [4,] 4 14
## [5,] 5 15
## [6,] 6 16
## [7,] 7 17
## [8,] 8 18
## [9,] 9 19
## [10,] 10 20Exercise: Create a matrix that contains the sequence of numbers from 1 to 16, going up in increments of 1. Let the matrix have 4 rows and have the matrix elements fill by row. Call this matrix M1.
To check the dimensions of a matrix, you can use the function dim().
## [1] 2 10## [1] 10 2Using square brackets [,] you can take subsets of matrices, the first element in the square bracket corresponds to the row indexing and the second element corresponds to the column indexing, [row, column].
## [1] 1 2 3 4## [1] 1 5 9 13You can subset multiple rows and multiple columns by indexing using vectors.
## [,1] [,2] [,3] [,4]
## [1,] 1 2 3 4
## [2,] 5 6 7 8## [,1] [,2]
## [1,] 3 4
## [2,] 7 8
## [3,] 11 12
## [4,] 15 16Both rows and columns can be indexed for selecting either a single element from a matrix or indexing a selection of rows and columns only.
## [1] 16#a matrix containing elements present in the first and second rows AND the
#third and fourth columns
mat[c(1,2), c(3,4)] ## [,1] [,2]
## [1,] 3 4
## [2,] 7 8Exercise: Extract the 3rd and 4th rows and the 1st and 2nd columns of your matrix, M1. Call this matrix M2.
As with vectors, through indexing, elements of the matrix can be updated.
citrus_mat <- matrix(data = c("oranges", "lemons", "limes", "apples"),
nrow = 2, ncol = 2, byrow = TRUE)
citrus_mat## [,1] [,2]
## [1,] "oranges" "lemons"
## [2,] "limes" "apples"#update the element in the second (2nd) row and the second (2nd) column to also
#be a citrus fruit
citrus_mat[2,2] <- "pomelo"
citrus_mat## [,1] [,2]
## [1,] "oranges" "lemons"
## [2,] "limes" "pomelo"The functions rowSums() and colSums() are used to find the row sums and column sums respectively, returning a vector of the values.
## [1] 10 26 42 58## [1] 28 32 36 40The apply() function can be used to obtain row or column summaries of a data matrix, with the following arguments:
X: the matrix you wish to summariseMARGIN: a vector that indicates which subscripts to apply the function to. For example,1indicates all rows,2indicates all columns andc(1,2)indicates rows and columnsFUN: the function that is to be applied to the datasimplify: logical, ifTRUE(the default), results will be simplified if possible
## [1] 28 32 36 40## [1] 2.5 6.5 10.5 14.5Exercise: Find the row sums of the matrix M2.
To multiply two matrices together, the operator %*% can be used, providing that the two matrices conform (the number of columns of one matrix must be equal to the number of rows on the other one).
## [,1] [,2]
## [1,] 4 4
## [2,] 4 41.4.3 Arrays
An array contains one or more matrices.
Create an array using the function array() in R, with the following arguments:
data: a vector containing data to fill the arraydim: an integer vector of length one or more giving the dimensions of the arraydimnames: alistwith one component for each dimension of the array
#an array of 3, 3x3 matrices
array1 <- array(c(round(seq(1,36, length=9)),
round(seq(10,30, length=9)),
round(seq(40,200, length=9))),
dim=c(nrow=3,ncol=3, 3))Arrays can be subset in the same way as matrices, using square brackets, except with an extra comma to indicate which of the matrices in the array is to be indexed.
## [,1] [,2] [,3]
## [1,] 1 14 27
## [2,] 5 18 32
## [3,] 10 23 361.4.4 Data frames
A data frame is a two-dimensional, tabular data type which can store multiple data types in R. It is also the most widely used data format as it can combine different types of variables.
You can create a data frame by using the function data.frame(), combining collections of variables.
For an example, a data frame can be constructed by creating some dummy/fake data:
##--Create some dummy or fake data
#paste0 function pastes everything together, returns region1, ..., region6
regions = paste0("region", 1:6)
tot_pop <- c(1000000, 920000, 2050000, 3100000, 1535000, 743000)
tot_hh <- c(200505, 124000, 882012,1051200, 452000, 79000)
male_prop <- c(0.55, 0.49, 0.45, 0.58, 0.56, 0.55)
male_pop <- tot_pop*male_prop
female_pop <- tot_pop - male_popand then using the function data.frame(), using the variables created above as the arguments:
data_frame1 <- data.frame(Region = regions,Tot_pop = tot_pop,Tot_hh = tot_hh,
Male_pop = male_pop, Female_pop = female_pop)
data_frame1## Region Tot_pop Tot_hh Male_pop Female_pop
## 1 region1 1000000 200505 550000 450000
## 2 region2 920000 124000 450800 469200
## 3 region3 2050000 882012 922500 1127500
## 4 region4 3100000 1051200 1798000 1302000
## 5 region5 1535000 452000 859600 675400
## 6 region6 743000 79000 408650 3343501.4.5 List
A list is a vector where each element itself is an object. Informally, it can be described as a ‘bag’ that contains data of different forms, types and dimensions, including other lists.
Create a list using the function list() in R, for example:
#create a list using the variables, matrices and arrays from above
list1 <- list(Admin = Regions, matrix = mat, array = array1)
list1## $Admin
## [1] "Northern" "Northeastern" "Western" "Central" "Eastern" "Southern"
##
## $matrix
## [,1] [,2] [,3] [,4]
## [1,] 1 2 3 4
## [2,] 5 6 7 8
## [3,] 9 10 11 12
## [4,] 13 14 15 16
##
## $array
## , , 1
##
## [,1] [,2] [,3]
## [1,] 1 14 27
## [2,] 5 18 32
## [3,] 10 23 36
##
## , , 2
##
## [,1] [,2] [,3]
## [1,] 10 18 25
## [2,] 12 20 28
## [3,] 15 22 30
##
## , , 3
##
## [,1] [,2] [,3]
## [1,] 40 100 160
## [2,] 60 120 180
## [3,] 80 140 200Data can be retrieved from a list using the dollar sign $, and subset the retrieved data in the same way as without lists.
## [1] "Northern" "Northeastern" "Western" "Central" "Eastern" "Southern"## NULLLogical operators can be used to subset data within a list.
## logical(0)## [,1] [,2] [,3] [,4]
## [1,] FALSE FALSE FALSE FALSE
## [2,] FALSE FALSE FALSE FALSE
## [3,] FALSE TRUE FALSE FALSE
## [4,] FALSE FALSE FALSE FALSEUsing the dollar sign, $, new variables can be added to a list.
## $Admin
## [1] "Northern" "Northeastern" "Western" "Central" "Eastern" "Southern"
##
## $matrix
## [,1] [,2] [,3] [,4]
## [1,] 1 2 3 4
## [2,] 5 6 7 8
## [3,] 9 10 11 12
## [4,] 13 14 15 16
##
## $array
## , , 1
##
## [,1] [,2] [,3]
## [1,] 1 14 27
## [2,] 5 18 32
## [3,] 10 23 36
##
## , , 2
##
## [,1] [,2] [,3]
## [1,] 10 18 25
## [2,] 12 20 28
## [3,] 15 22 30
##
## , , 3
##
## [,1] [,2] [,3]
## [1,] 40 100 160
## [2,] 60 120 180
## [3,] 80 140 200
##
##
## $vector
## [1] 10 20 30 40 50 60 70 80 90 100 15 15 14 14 13 13 12 12 11 11 10 10Lists can be also be created to demonstrate different levels, for example, age ranges.
Age <- list(Infant = "0-59 months",
Child = "5 - 12 years",
Teen = "13 - 17 years",
Adult = "18+")
Age## $Infant
## [1] "0-59 months"
##
## $Child
## [1] "5 - 12 years"
##
## $Teen
## [1] "13 - 17 years"
##
## $Adult
## [1] "18+"Lists can be ‘flattened’ using the function unlist(), which simplifies the list into a vector which contains the same elements.
## Infant Child Teen Adult
## "0-59 months" "5 - 12 years" "13 - 17 years" "18+"1.5 End of module exercises
1. Use R to find the value of the square root of 25. (\(\sqrt(25)\))
Hint: Look at the help file for the function sqrt().
2. Use R to find the value of the exponential of \(6\times 14 -3\). (\(\exp(6\times 14-3)\))
3. Use R to find the value of the absolute value of \(7\times 3 - 4\times 9 - 30\). (\(|7\times 3 - 4\times 9 - 30|\))
4. Use R to find the value of \(\frac{73-42}{3}+2\times\left(\frac{36}{4}-17\right)\). Give your answer to 2 decimal places.
5. How many elements does the following vector have?
seq(from = 0, to = 100, by = 3.14)
6. Look at the help file for the function rnorm() and use this function to generate 100 random numbers with mean 10 and standard deviation 5.