The foot package was developed by WorldPop at the University of Southampton (www.worldpop.org) to support geometric calculations and zonal summaries of measures from building footprint polygons. The vignette("footsteps", package="foot") provides an introduction to package and the functionality of calculating and summarising morphology measures. This vignette builds on those methods and demonstrates a more advanced workflow to produce gridded summaries of buildings measures. In particular, the focus is on using foot::calculate_bigfoot() in order how to handle very large (i.e. national-scale) datasets of building footprints.
An example of this type of processing and how it can be used in settlement classification is available in this paper (open access): https://doi.org/10.1371/journal.pone.0247535.
Options and finer control
The calculate_bigfoot function is set up with default values that should work under most conditions; however, there is additional flexibility for users to specify alternative parameters.
Specifying geometry units
To override the default units used in the geometry calculations, a named list of unit strings can be supplied to the controlUnits argument. This list can contain named items for areaUnit, perimUnit, and distUnit. The default values are meters (“m”) and square meters (“m^2”) The value of each item should be coercible with units::as_units.
# change the default units used to calculate area and distance
calculate_bigfoot(X=buildings,
template=mgrid,
what=list(list("area"), list("perimeter")),
how=list(list("mean"), list("sum")),
controlZone=list("method"="intersect"), # join by intersection
controlUnits=list(areaUnit="m^2", # change default units
perimUnit="km"),
outputPath=tempdir(),
outputTag="TAG",
parallel=FALSE,
verbose=TRUE)
#> Reading template grid
#> trying to read file: C:\Users\Admin\Documents\GitHub\foot\wd\in\kampala_grid.tif
#> Selecting metrics
#> Setting control values
#> Creating output grids
#> Creating list of processing tiles
#>
#> Tile: 1 of 1
#> Reading footprints
#> Reading template grid
#> Selecting metrics
#> Setting control values.
#> Pre-calculating areas
#> Pre-calculating perimeters
#> Creating zonal index
#>
#> Calculating 2 metrics ...
#> area mean
#> perimeter sum
#> Finished calculating metrics.
#> Writing output tiles
#> Finished writing grids
#>
#> Finished processing all tiles: 2021-11-10 18:02:12
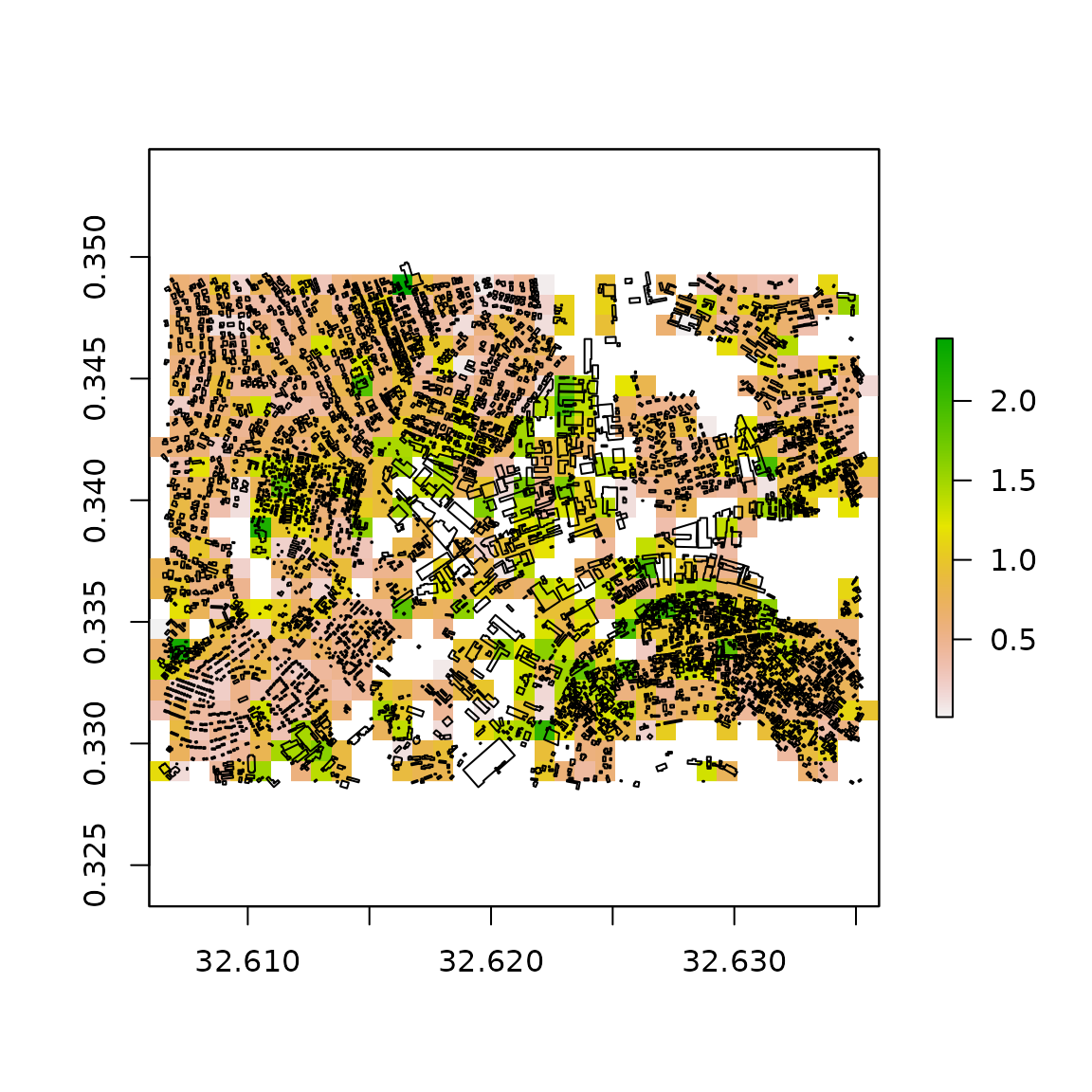
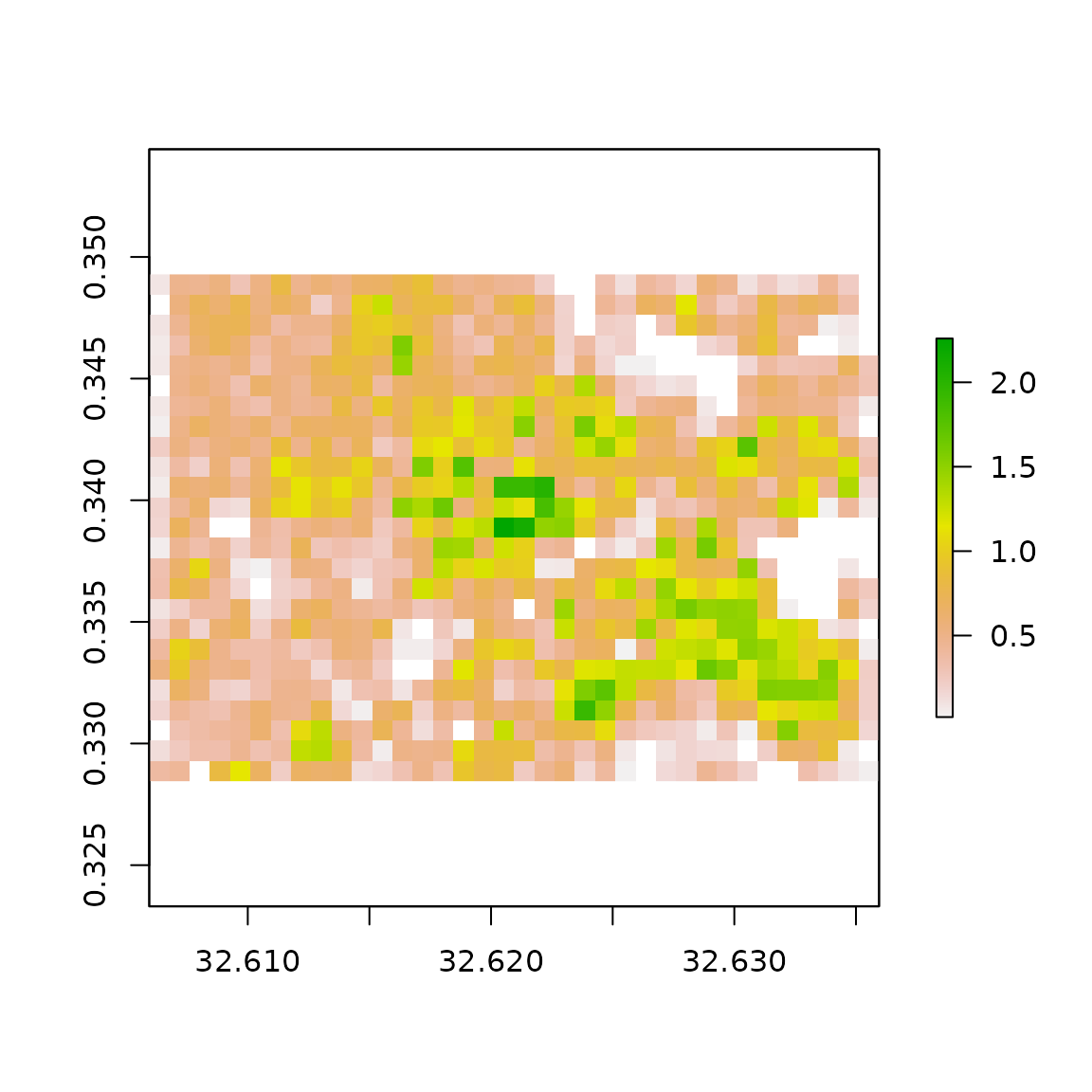
# plot the total perimeter, measured in kilometres
outGrid <- raster::raster(file.path(tempdir(), "TAG_perimeter_sum.tif"))
raster::plot(outGrid)
Filtering buildings
In some settings it may be preferable to exclude very small and/or very large building footprint polygons. The lower and upper bounds for filtering can be specified with minArea and maxArea in the filter argument. The values for these filters are in the same units specified by controlUnits or the default value for area calculations. Note that an “area” footprint statistic does not need to be requested as this characteristic is automatically calculated to enable filtering.
# Filtering: # footprints must be larger than 50 m^2 and smaller than 1000 m^2
calculate_bigfoot(X=buildings,
template=mgrid,
what=list(list("shape"), list("settled"), list("perimeter")),
how=list(list("mean"), list("count"), list("sum")),
controlUnits=list(areaUnit="m^2"),
filter=list(minArea=50, maxArea=1000),
outputPath=tempdir(),
parallel=FALSE,
verbose=TRUE)
#> Reading template grid
#> trying to read file: C:\Users\Admin\Documents\GitHub\foot\wd\in\kampala_grid.tif
#> Selecting metrics
#> Setting control values
#> Creating output grids
#> Creating list of processing tiles
#>
#> Tile: 1 of 1
#> Reading footprints
#> Reading template grid
#> Selecting metrics
#> Setting control values.
#> Pre-calculating areas
#> Pre-calculating perimeters
#> Pre-calculating shape
#> Creating zonal index
#> Filtering features larger than 50
#> Filtering features smaller than 1000
#>
#> Calculating 3 metrics ...
#> shape mean
#> settled count
#> perimeter sum
#> Finished calculating metrics.
#> Writing output tiles
#> Finished writing grids
#>
#> Finished processing all tiles: 2021-11-10 18:02:20
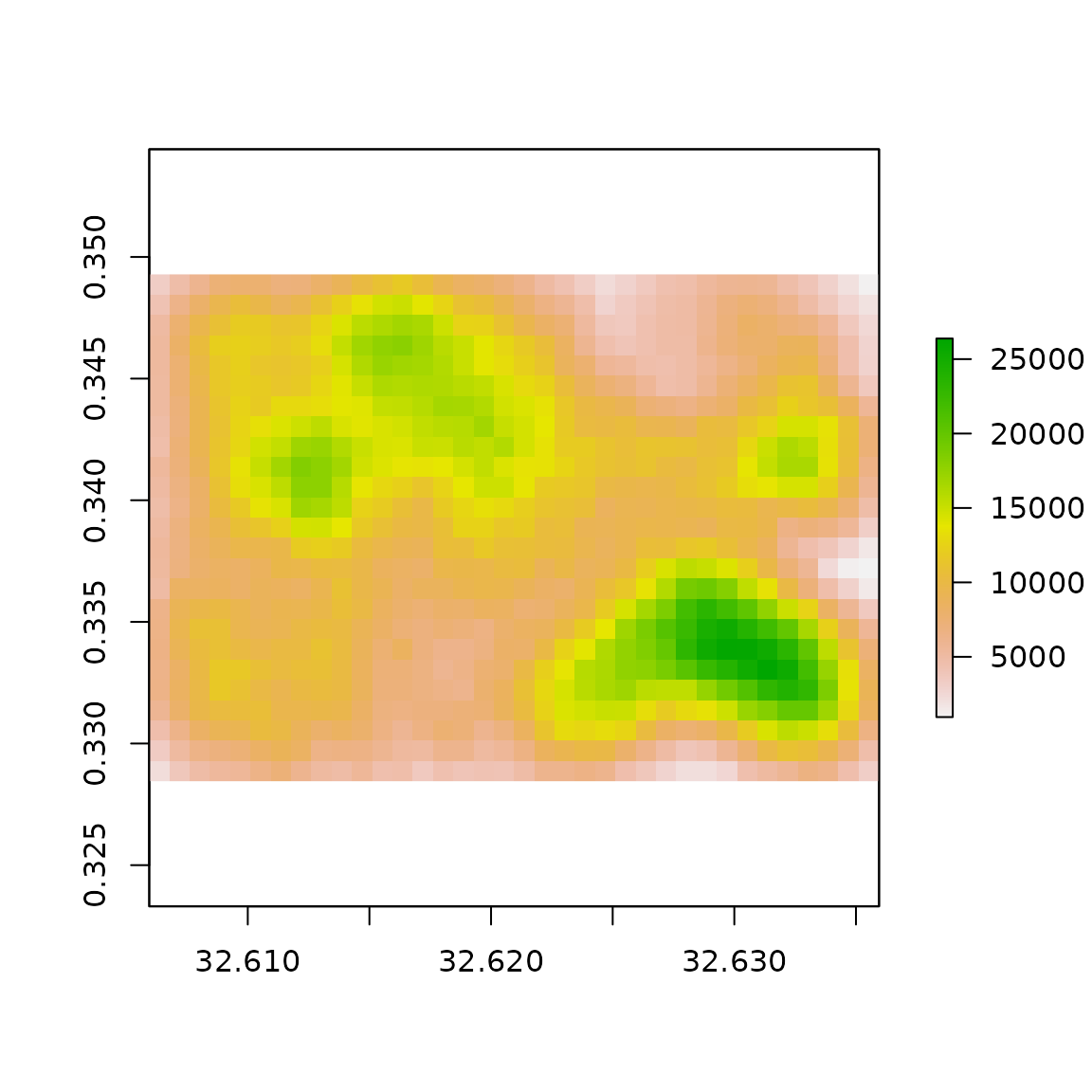
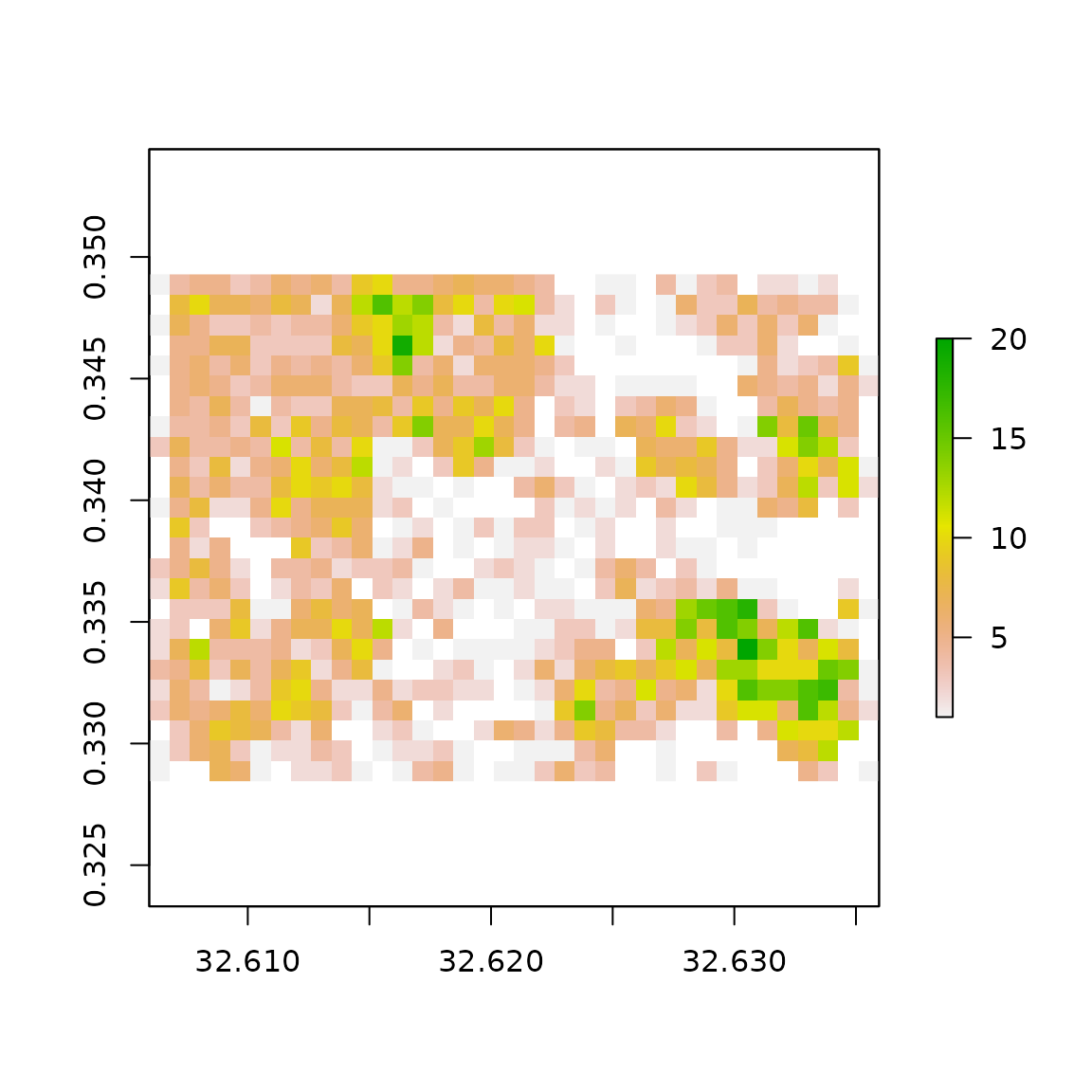
In the map of the results below, each pixel is the count of footprints present. Note the smaller number of structures and sparseness of structures in pixels around the centre portions of the image. This corresponds with the a business district and industrial areas with fewer, but larger structures.
Tile size
The size of the processing tiles, specified in pixel dimensions (rows, columns) can be an important factor in the efficiency of the calculations. Smaller tile regions result in fewer building footprints being read/processed at one time, but there is an overhead computational cost of reading/writing files. The default value is 500 pixels. For the small demonstration shown here that results in one tile for the whole region. To show multiple tile processing, a small size is supplied and the processing is done in parallel with verbose output.
calculate_bigfoot(X=buildings,
template=mgrid,
what="compact",
how="mean",
tileSize=c(20, 20), # rows x columns in pixels
parallel=FALSE,
verbose=TRUE)
#> Reading template grid
#> trying to read file: C:\Users\Admin\Documents\GitHub\foot\wd\in\kampala_grid.tif
#> Selecting metrics
#> Setting control values
#> Creating output grids
#> Creating list of processing tiles
#>
#> Tile: 1 of 4
#> Reading footprints
#> Reading template grid
#> Selecting metrics
#> Setting control values.
#> Pre-calculating compactness
#> Creating zonal index
#>
#> Calculating 1 metrics ...
#> compact mean
#> Finished calculating metrics.
#> Writing output tiles
#> Finished writing grids
#>
#> Tile: 2 of 4
#> Reading footprints
#> Reading template grid
#> Selecting metrics
#> Setting control values.
#> Pre-calculating compactness
#> Creating zonal index
#>
#> Calculating 1 metrics ...
#> compact mean
#> Finished calculating metrics.
#> Writing output tiles
#> Finished writing grids
#>
#> Tile: 3 of 4
#> Reading footprints
#> Reading template grid
#> Selecting metrics
#> Setting control values.
#> Pre-calculating compactness
#> Creating zonal index
#>
#> Calculating 1 metrics ...
#> compact mean
#> Finished calculating metrics.
#> Writing output tiles
#> Finished writing grids
#>
#> Tile: 4 of 4
#> Reading footprints
#> Reading template grid
#> Selecting metrics
#> Setting control values.
#> Pre-calculating compactness
#> Creating zonal index
#>
#> Calculating 1 metrics ...
#> compact mean
#> Finished calculating metrics.
#> Writing output tiles
#> Finished writing grids
#>
#> Finished processing all tiles: 2021-11-10 18:02:27
Restarting
When running parallel processes on clusters, it is not currently possible to show a progress bar. This issue will hopefully be addressed in the future. However, when verbose=TRUE, bigfoot will write a log file to the output directory with the gridded data. This file is a list of tile ID numbers that have been processed. Tiles (created with gridTiles) are numbered sequentially from top-left to bottom-right of the template grid. On long-running jobs, the log of completed tiles can be used to monitor progress. Note that each processor will write its results as they are finished, so tiles may not be completed in order.
If a processing job crashes, the log file can also be used to restart the job. Setting the argument restart to a number will restart processing from that tile. Alternatively a sequence of numbers, given as a vector, can be used to re-run only certain tiles. The output grids will not be overwritten when restarting.
Next steps
This vignette has provided an overview of how to create gridded outputs layers summarising building footprint morphology measures. This workflow uses calculate_bigfoot and is designed to work for large sets of data through tiled read/writing and processing these tiles in parallel. The bigfoot functionality extends and makes use of footstats. Both of these functions can take use-supplied and custom summary functions. This advanced usage is demonstrated in vignette("cobbler", package="foot").
sessionInfo()
#> R version 4.1.1 (2021-08-10)
#> Platform: x86_64-pc-linux-gnu (64-bit)
#> Running under: Ubuntu 20.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
#> LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
#>
#> locale:
#> [1] LC_CTYPE=en_GB.UTF-8 LC_NUMERIC=C
#> [3] LC_TIME=en_GB.UTF-8 LC_COLLATE=en_GB.UTF-8
#> [5] LC_MONETARY=en_GB.UTF-8 LC_MESSAGES=en_GB.UTF-8
#> [7] LC_PAPER=en_GB.UTF-8 LC_NAME=C
#> [9] LC_ADDRESS=C LC_TELEPHONE=C
#> [11] LC_MEASUREMENT=en_GB.UTF-8 LC_IDENTIFICATION=C
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] sf_1.0-0 raster_3.4-5 sp_1.4-5 foot_0.8
#>
#> loaded via a namespace (and not attached):
#> [1] Rcpp_1.0.7 lattice_0.20-45 class_7.3-19 rprojroot_2.0.2
#> [5] digest_0.6.28 foreach_1.5.1 utf8_1.2.2 R6_2.5.1
#> [9] evaluate_0.14 e1071_1.7-7 highr_0.9 pillar_1.6.4
#> [13] rlang_0.4.12 data.table_1.14.2 jquerylib_0.1.3 rmarkdown_2.11
#> [17] pkgdown_1.6.1 textshaping_0.3.5 desc_1.4.0 rgdal_1.5-23
#> [21] stringr_1.4.0 proxy_0.4-26 compiler_4.1.1 xfun_0.27
#> [25] pkgconfig_2.0.3 systemfonts_1.0.3 htmltools_0.5.2 tidyselect_1.1.0
#> [29] tibble_3.1.5 codetools_0.2-18 fansi_0.5.0 crayon_1.4.2
#> [33] dplyr_1.0.5 wk_0.4.1 grid_4.1.1 jsonlite_1.7.2
#> [37] lwgeom_0.2-6 lifecycle_1.0.1 DBI_1.1.1 magrittr_2.0.1
#> [41] formatR_1.11 units_0.7-2 KernSmooth_2.23-20 stringi_1.7.5
#> [45] cachem_1.0.6 fs_1.5.0 doParallel_1.0.16 bslib_0.2.4
#> [49] ellipsis_0.3.2 filelock_1.0.2 ragg_1.1.3 generics_0.1.0
#> [53] vctrs_0.3.8 s2_1.0.6 iterators_1.0.13 tools_4.1.1
#> [57] glue_1.4.2 purrr_0.3.4 abind_1.4-5 parallel_4.1.1
#> [61] fastmap_1.1.0 yaml_2.2.1 stars_0.5-3 classInt_0.4-3
#> [65] memoise_2.0.0 knitr_1.36 sass_0.3.1